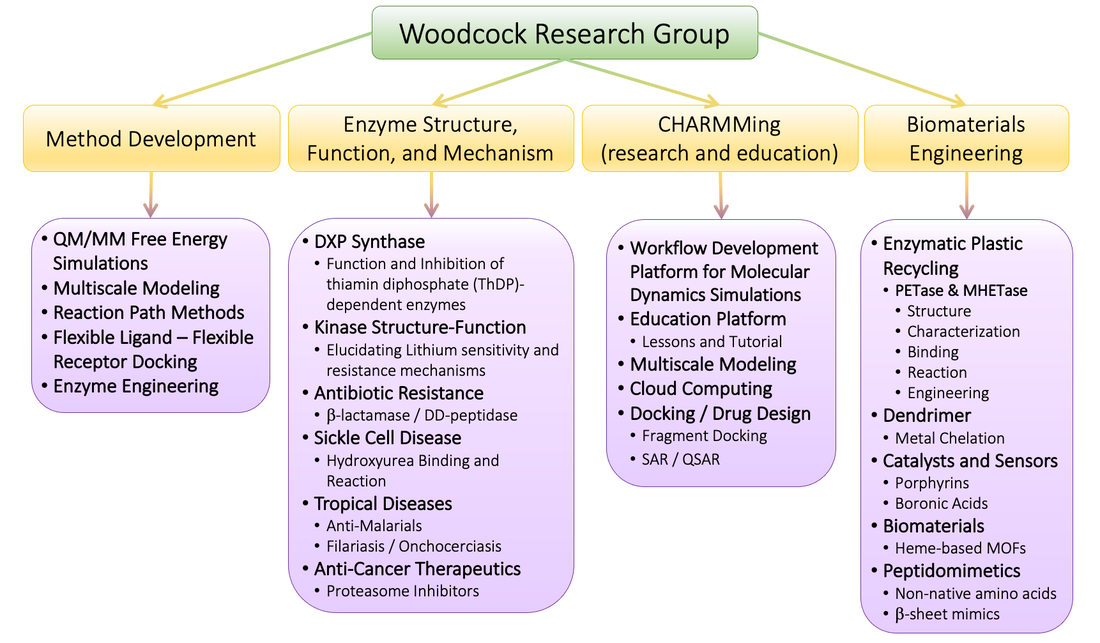
My current and future research will primarily focus on developing and employing computational methodology to solve interesting problems that exist at the interface of biophysics, medicine, and materials (see below for a visual overview). More information can be found on ORCID: https://orcid.org/0000-0003-3539-273
Funded Research
Design and application of robust and efficient QM/MM free energy simulation methods for biomolecular systems (PI: R01GM129519).
To accurately compute free energies of complex biomolecular systems there are two key prerequisites: the accurate description of inter- and intramolecular interactions, and adequate sampling of all relevant conformational degrees of freedom. Include the possibility that conformational dynamics may be coupled to complex electronic processes or chemical reactions, where quantum mechanical (QM) methods are needed, and this task becomes extremely daunting. Currently, whenever accurate computations of biomolecular systems are essential, the tool of choice is hybrid quantum mechanical / molecular mechanical (QM/MM) calculations, however, the application of these techniques to free energy simulations (FES) is far from common. Herein, we aim to develop a set of robust, efficient, and accurate new techniques that will make the application of QM/MM FES nearly routine. These methods will be subsequently applied to study two classes of biomolecular applications that present extreme challenges to current techniques. Namely, the accurate computation of (1) absolute pKa as a function of ligand binding and reaction (e.g., accounting for transient interactions), and (2) free energy differences of binding as a function of tautomerism. The scientific results will be closely tied to activities aimed at improving access to these advanced techniques.
Toward understanding the chemistry and biology of microbial DXP synthase (co-PI: R01GM143810).
Microbial metabolic function and adaptability are important in many facets of human health and disease. This proposal centers on studies of the microbial metabolic enzyme 1-deoxy-D-xylulose 5-phosphate synthase (DXPS). DXPS catalyzes the thiamin diphosphate (ThDP)-dependent synthesis of the essential metabolite DXP, which is found in the gut microbiota, pathogenic bacteria and parasites, but not in humans. Positioned at a metabolic branchpoint, DXP serves as a precursor to indispensable isoprenoids and vitamins, thus, we postulate DXPS plays key roles in microbiome function and microbial metabolic adaptation. Our long-term goal is to learn how microbes use DXPS in different contexts, toward understanding its roles in the microbiome and its potential as an antimicrobial target. Our group discovered a unique ligand-gated mechanism used by DXPS that bestows targets for selective inhibition and may enable DXPS to sense and respond to its environment. The goals of this proposal are to dissect how DXPS responds to molecular cues, examine this sensing mechanism in different chemical contexts, and establish novel inhibition strategies that target this distinctive feature of DXPS. Insights gained from this research will direct our thinking about DXPS function, and guide development of chemical probes needed to study DXPS roles in microbes.
Data Management for Molecule Simulation : A Throughput-Oriented Approach (co-I: R01GM140316).
Project Summary We will design and develop a molecular simulation data management system, we call P2DMS, to analyze large distributed molecular dynamics (MD) simulation data. The salient features of the system include: (1) a push-based local query engine design that handles data in a batch processing manner and processes many queries at the same time; (2) optimized MD analytics tools using modern many-core hardware such as GPUs; and (3) efficient management and access to distributed data over wide area networks, which is quite common for large scale MD simulations. This will be done by building a data analysis layer on top of state-of-the-art distributed big data management systems. The out- come of this project will not only improve the efficiency of MD data processing, but also enable new knowledge discovery that is currently regarded difficult or infeasible. In particular, we will integrate the P2DMS program into existing MD simulation packages, and validate the new design with important real-world biological and MD methodological problems. In particular, we will (1) model the structure-function relationships of how the spike protein of SARS-CoV-2 inter- acts with the human angiotensin converting enzyme 2 (ACE2) receptor; and (2) enhance the performance of a recently developed parameter optimization software for active control of MD simulations.
To accurately compute free energies of complex biomolecular systems there are two key prerequisites: the accurate description of inter- and intramolecular interactions, and adequate sampling of all relevant conformational degrees of freedom. Include the possibility that conformational dynamics may be coupled to complex electronic processes or chemical reactions, where quantum mechanical (QM) methods are needed, and this task becomes extremely daunting. Currently, whenever accurate computations of biomolecular systems are essential, the tool of choice is hybrid quantum mechanical / molecular mechanical (QM/MM) calculations, however, the application of these techniques to free energy simulations (FES) is far from common. Herein, we aim to develop a set of robust, efficient, and accurate new techniques that will make the application of QM/MM FES nearly routine. These methods will be subsequently applied to study two classes of biomolecular applications that present extreme challenges to current techniques. Namely, the accurate computation of (1) absolute pKa as a function of ligand binding and reaction (e.g., accounting for transient interactions), and (2) free energy differences of binding as a function of tautomerism. The scientific results will be closely tied to activities aimed at improving access to these advanced techniques.
Toward understanding the chemistry and biology of microbial DXP synthase (co-PI: R01GM143810).
Microbial metabolic function and adaptability are important in many facets of human health and disease. This proposal centers on studies of the microbial metabolic enzyme 1-deoxy-D-xylulose 5-phosphate synthase (DXPS). DXPS catalyzes the thiamin diphosphate (ThDP)-dependent synthesis of the essential metabolite DXP, which is found in the gut microbiota, pathogenic bacteria and parasites, but not in humans. Positioned at a metabolic branchpoint, DXP serves as a precursor to indispensable isoprenoids and vitamins, thus, we postulate DXPS plays key roles in microbiome function and microbial metabolic adaptation. Our long-term goal is to learn how microbes use DXPS in different contexts, toward understanding its roles in the microbiome and its potential as an antimicrobial target. Our group discovered a unique ligand-gated mechanism used by DXPS that bestows targets for selective inhibition and may enable DXPS to sense and respond to its environment. The goals of this proposal are to dissect how DXPS responds to molecular cues, examine this sensing mechanism in different chemical contexts, and establish novel inhibition strategies that target this distinctive feature of DXPS. Insights gained from this research will direct our thinking about DXPS function, and guide development of chemical probes needed to study DXPS roles in microbes.
Data Management for Molecule Simulation : A Throughput-Oriented Approach (co-I: R01GM140316).
Project Summary We will design and develop a molecular simulation data management system, we call P2DMS, to analyze large distributed molecular dynamics (MD) simulation data. The salient features of the system include: (1) a push-based local query engine design that handles data in a batch processing manner and processes many queries at the same time; (2) optimized MD analytics tools using modern many-core hardware such as GPUs; and (3) efficient management and access to distributed data over wide area networks, which is quite common for large scale MD simulations. This will be done by building a data analysis layer on top of state-of-the-art distributed big data management systems. The out- come of this project will not only improve the efficiency of MD data processing, but also enable new knowledge discovery that is currently regarded difficult or infeasible. In particular, we will integrate the P2DMS program into existing MD simulation packages, and validate the new design with important real-world biological and MD methodological problems. In particular, we will (1) model the structure-function relationships of how the spike protein of SARS-CoV-2 inter- acts with the human angiotensin converting enzyme 2 (ACE2) receptor; and (2) enhance the performance of a recently developed parameter optimization software for active control of MD simulations.