|
|
B. R. Brooks, C. L. Brooks III, A. D. MacKerell, Jr., L. Nilsson, B. Roux, Y. Won, G. Archontis, C. Bartels, S. Boresch, L. Caves, Q. Cui, A. Dinner, S. Fischer, J. Gao, M. Hodoscek, K. Kuczera, T. Lazaridis, J. Ma, E. Paci, R. W. Pastor, C. B. Post, M. Schaefer, B. Tidor, R. M. Venable, H. L. Woodcock, X. Wu and M. Karplus; CHARMM The Biomolecular Simulation Program. J. Comp. Chem.; 2009; 30 (10); 1545-1614.
Q-Chem is a comprehensive ab initio quantum chemistry package. Its capabilities range from the highest performance DFT/HF calculations to high level post-HF correlation methods. Q-Chem tackles a wide range of problems in commercial, academic and government laboratories, including:
Y. Shao, L. Fusti-Molnar, Y. Jung, J. Kussmann, C. Ochsenfeld, S. T. Brown, A. T. B. Gilbert, L. V. lipchenko, S. V. Levchenko, D. P. ONeill, R. A. Distasio, R. C. Lochan, T. Wang, G. J. O. Beran, N. A. Besley, J. M. Herbert, C. Y. Lin, T. Van Voorhis, S. H. Chien, A. Sodt, R. P. Steele, V. A. Rassolov, P. E. Maslen, P. P. Korambath, R. D. Adamson, B. Austin, J. Baker, E. F. C. Byrd, H. Daschel, R. J. Doerksen, A. Dreuw, B. D. Dunietz, A. D. Dutoi, T. R. Furlani, S. R. Gwaltney, A. Heyden, S. Hirata, C. P. Hsu, G. Kedziora, R. Z. Khalliulin, P. Klunzinger, A. M. Lee, M. S. Lee, W. Liang, I. Lotan, N. Nair, B. Peters, E. I. Proynov, P. A. Pieniazek, Y. M. Rhee, J. Ritchie, E. Rosta, D. C. Sherrill, A. C. Simmonett, J. E. Subotnik, H. Lee Woodcock, W. Zhang, A. T. Bell, A. K. Chakraborty, D. M. Chipman, F. J. Keil, A. Warshel, W. J. Hehre, H. F. Schaefer, J. Kong, A. I. Krylov, P. M. W. Gill, M. Head-Gordon; Advances in methods and algorithms in a modern quantum chemistry program package. (Phys. Chem. Chem. Phys.); 2006; 8; 3172-3191.
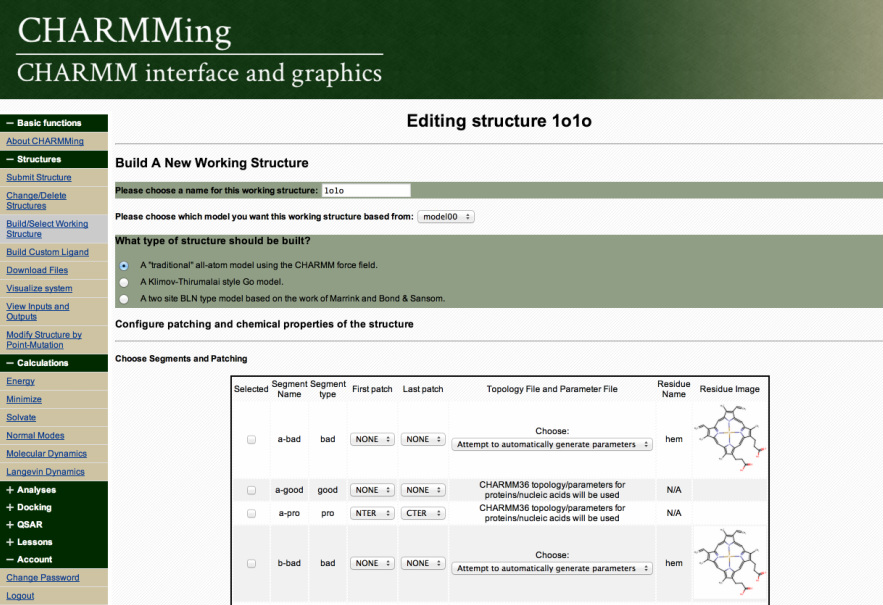
CHARMMing is a recently developed Web user interface for the CHARMM macromolecular simulation package.** This open- source tool provides a user-friendly interface for the preparation, submission, monitoring, and visualization of molecular simulations (i.e. energy minimization, solvation, and dynamics).
CHARMMing is divided into “applications”, each of which encapsulate some important part of the program. CHARMMing’s primary design considerations are flexibility, transparency, and adaptability. The software is built on top of the freely available Django web application framework, which handles various low-level details of web programming so that developers can concentrate on implementing functionality. Django was chosen for its technical capabilities and because it is built around the popular Python programming language.
CHARMMing is a modern software package (i.e. adhering to HTML5 standards, requiring no external Web plugins, etc.) and can be used with an up-to-date browser (e.g. Google Chrome, Firefox, Safari, etc.). Each application has its own data model which stores information about the particular task being performed and a set of functions which process commands and are collectively called the applications views. Data generated by one CHARMMing application can easily be used in another. For example, a structure can be energy minimized and then used to setup a MD simulation. The end result of this architecture is a highly flexible and easy-to-modify code.
** B. T. Miller, R. P. Singh, J. B. Klauda, M. Hodoscek, B. R. Brooks, and H. Lee Woodcock*; CHARMMing: A New, Flexible, Web-based front-end to CHARMM. J. Chem. Inf. Mod.; 2008; 48(9); 1920-1929.
CHARMMing is divided into “applications”, each of which encapsulate some important part of the program. CHARMMing’s primary design considerations are flexibility, transparency, and adaptability. The software is built on top of the freely available Django web application framework, which handles various low-level details of web programming so that developers can concentrate on implementing functionality. Django was chosen for its technical capabilities and because it is built around the popular Python programming language.
CHARMMing is a modern software package (i.e. adhering to HTML5 standards, requiring no external Web plugins, etc.) and can be used with an up-to-date browser (e.g. Google Chrome, Firefox, Safari, etc.). Each application has its own data model which stores information about the particular task being performed and a set of functions which process commands and are collectively called the applications views. Data generated by one CHARMMing application can easily be used in another. For example, a structure can be energy minimized and then used to setup a MD simulation. The end result of this architecture is a highly flexible and easy-to-modify code.
** B. T. Miller, R. P. Singh, J. B. Klauda, M. Hodoscek, B. R. Brooks, and H. Lee Woodcock*; CHARMMing: A New, Flexible, Web-based front-end to CHARMM. J. Chem. Inf. Mod.; 2008; 48(9); 1920-1929.
|
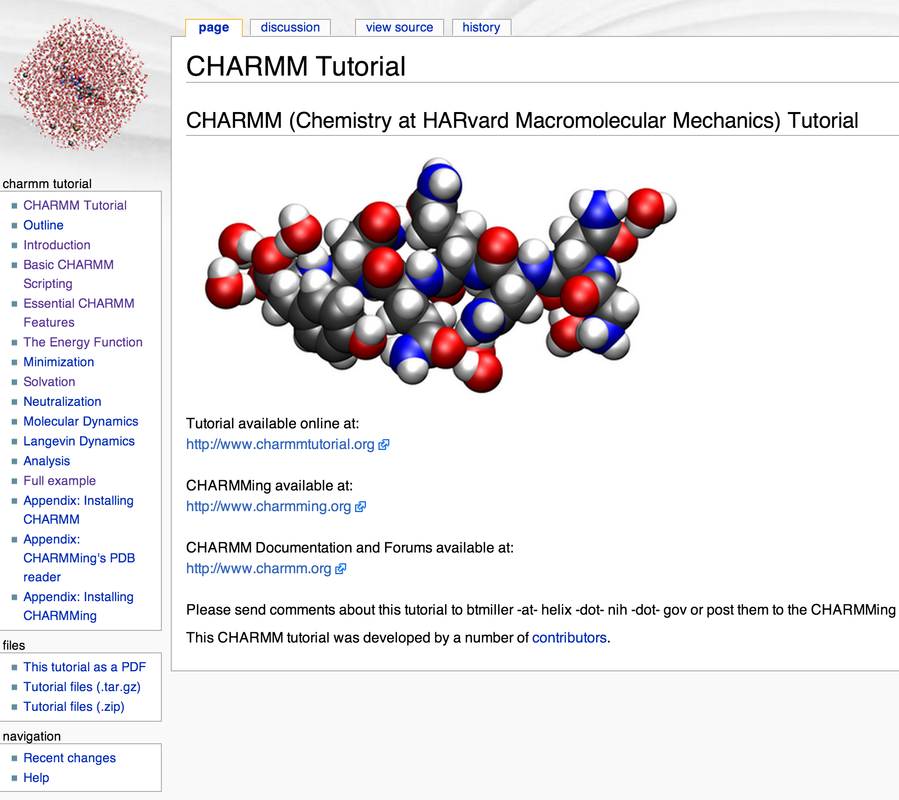
A tutorial/introduction to the use of CHARMM has been created and published on the web (CHARMM Tutorial).** In addition, this has been integrated into CHARMMing. This tutorial provides conceptual foundations to molecular simulation techniques such as energy minimization, solvation, molecular dynamics, and the use of periodic boundary conditions that are implemented in CHARMMing. Thus CHARMMing may be used as an introduction to molecular simulations in general and CHARMM in particular with the tutorial serving as a manual.
** Benjamin T. Miller, Rishi P. Singh, Vinushka Schalk, Stefan Boresch, Yuri Pevzner, Jingjun Sun, Carrie S. Miller, Toshiko Ichiye, Bernard R. Brooks, H. Lee Woodcock III*; Web based computational chemistry education with CHARMMing I: Lessons and Tutorial. PLoS Comput. Biol.; 2014; Submitted. |